Mathematical Modeling of Glycosylation Pathways
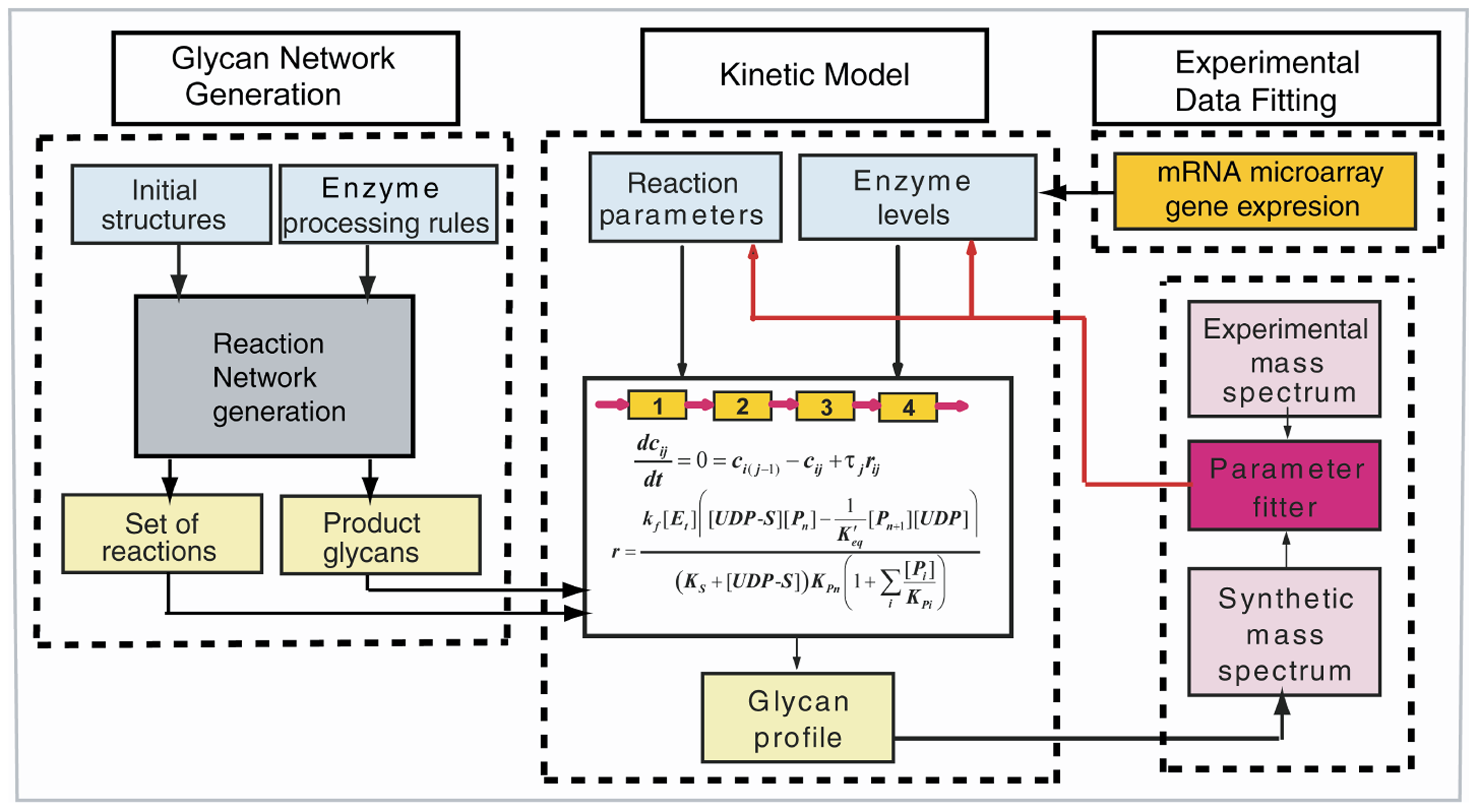
Glycans are the sugar attachments that are found on proteins and lipids. These highly variable and structurally diverse sugar chains confer distinctive characteristics to the cell surface. Recent research has revealed that abnormalities in glycan biosynthesis have been conclusively linked to many diseases, but the complexity of glycosylation has hindered the analysis of glycan data in order to identify glycoforms contributing to disease. To overcome this limitation, we developed a quantitative N-glycosylation model that interprets and integrates mass spectral and transcriptomic data by incorporating key glycosylation enzyme activities.
Using the cancer progression model of androgen-dependent to androgen-independent Lymph Node Carcinoma of the Prostate (LNCaP) cells, the N-glycosylation model identified and quantified glycan structural details not typically derived from single-stage mass spectral or gene expression data. Differences between the cell types uncovered include increases in H(II) and Ley epitopes, corresponding to greater activity of α2-Fuc-transferase (FUT1) in the androgen-independent cells. The model further elucidated limitations in the two analytical platforms including a defect in the microarray for detecting the GnTV (MGAT5) enzyme. Our results demonstrate the potential of systems glycobiology tools for elucidating key glycan biomarkers and potential therapeutic targets. The integration of multiple data sets represents an important application of systems biology for understanding complex cellular processes.
References
Krambeck, Frederick J., Sandra V. Bennun, Someet Narang, Sean Choi, Kevin J. Yarema, and Michael J. Betenbaugh. "A Mathematical Model to Derive N-Glycan Structures and Cellular Enzyme Activities from Mass Spectrometric Data." Glycobiology 19, no. 11 (November 01, 2009): 1163-1175.