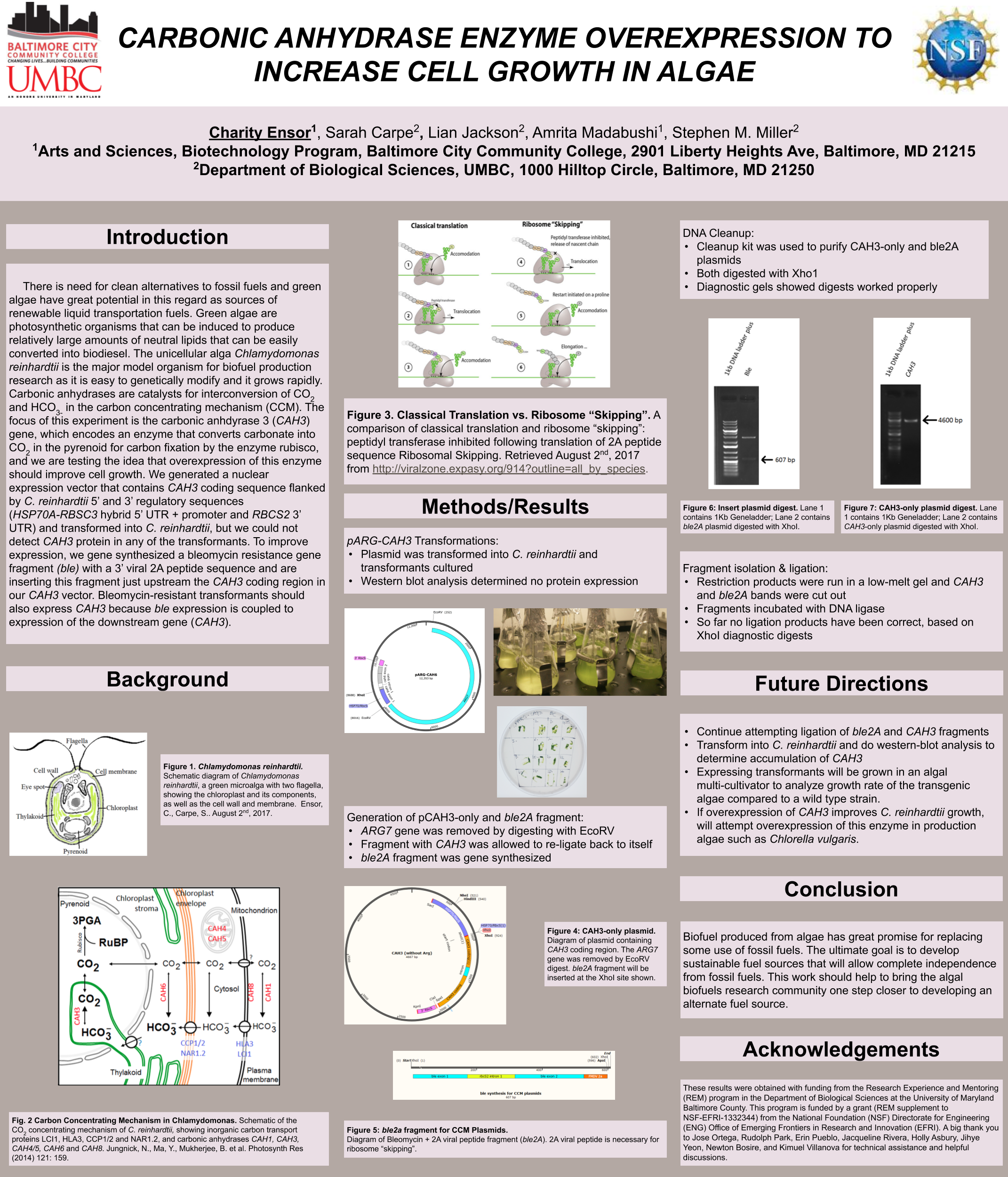
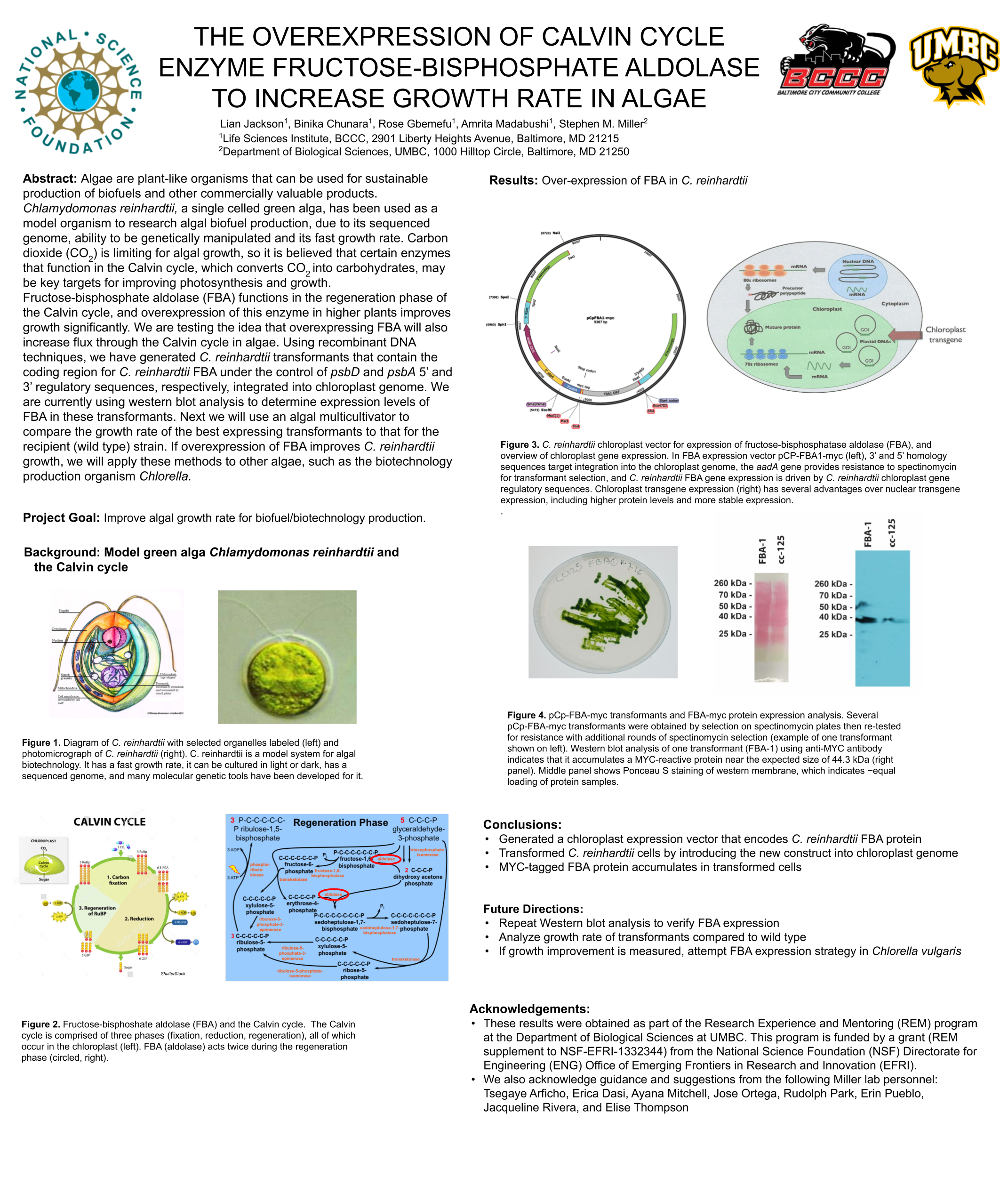
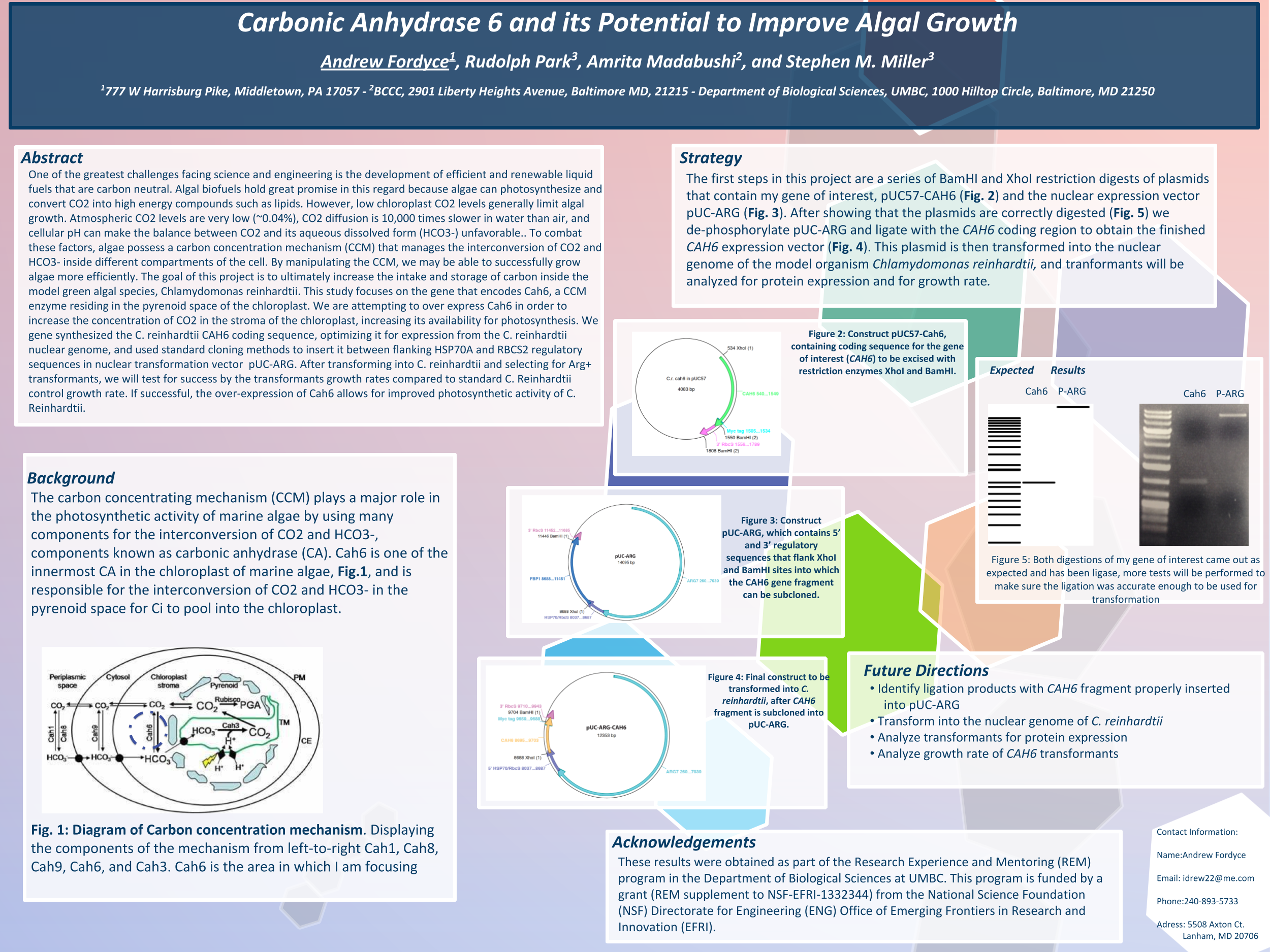
Towanda Carr, Yihun Asires, Amrita Madabushi, Lian Jackson, Stephen M. Miller
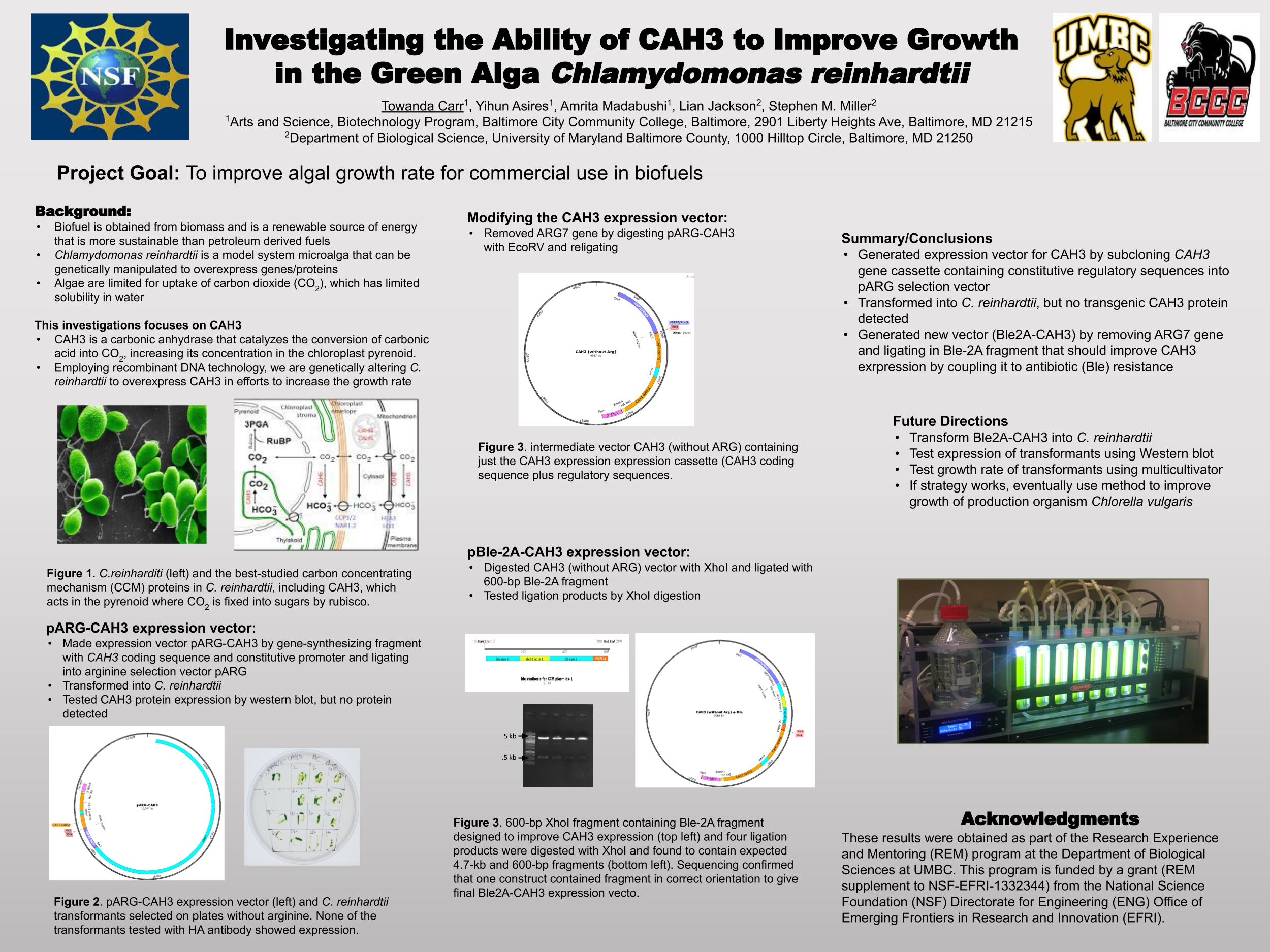
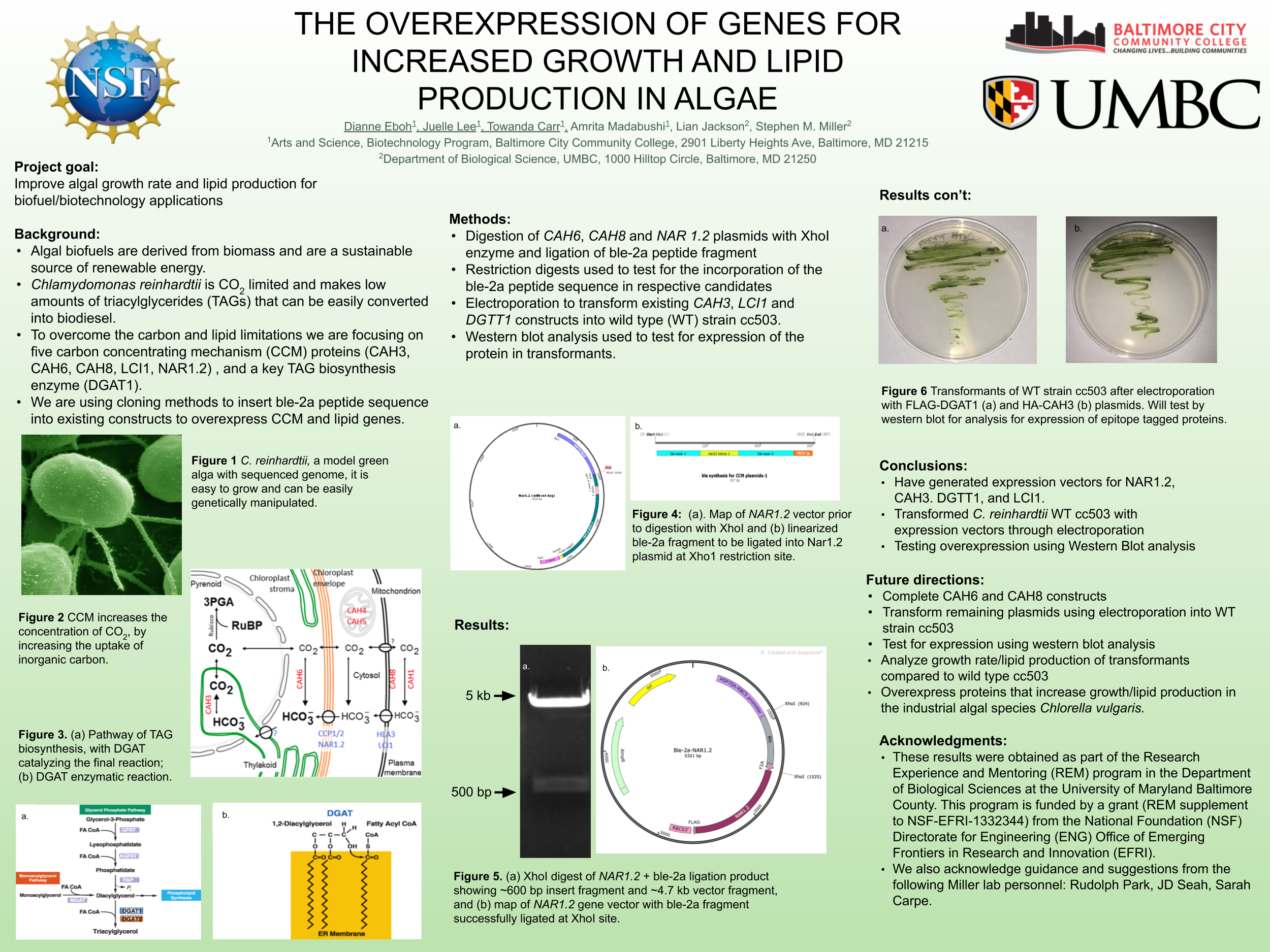
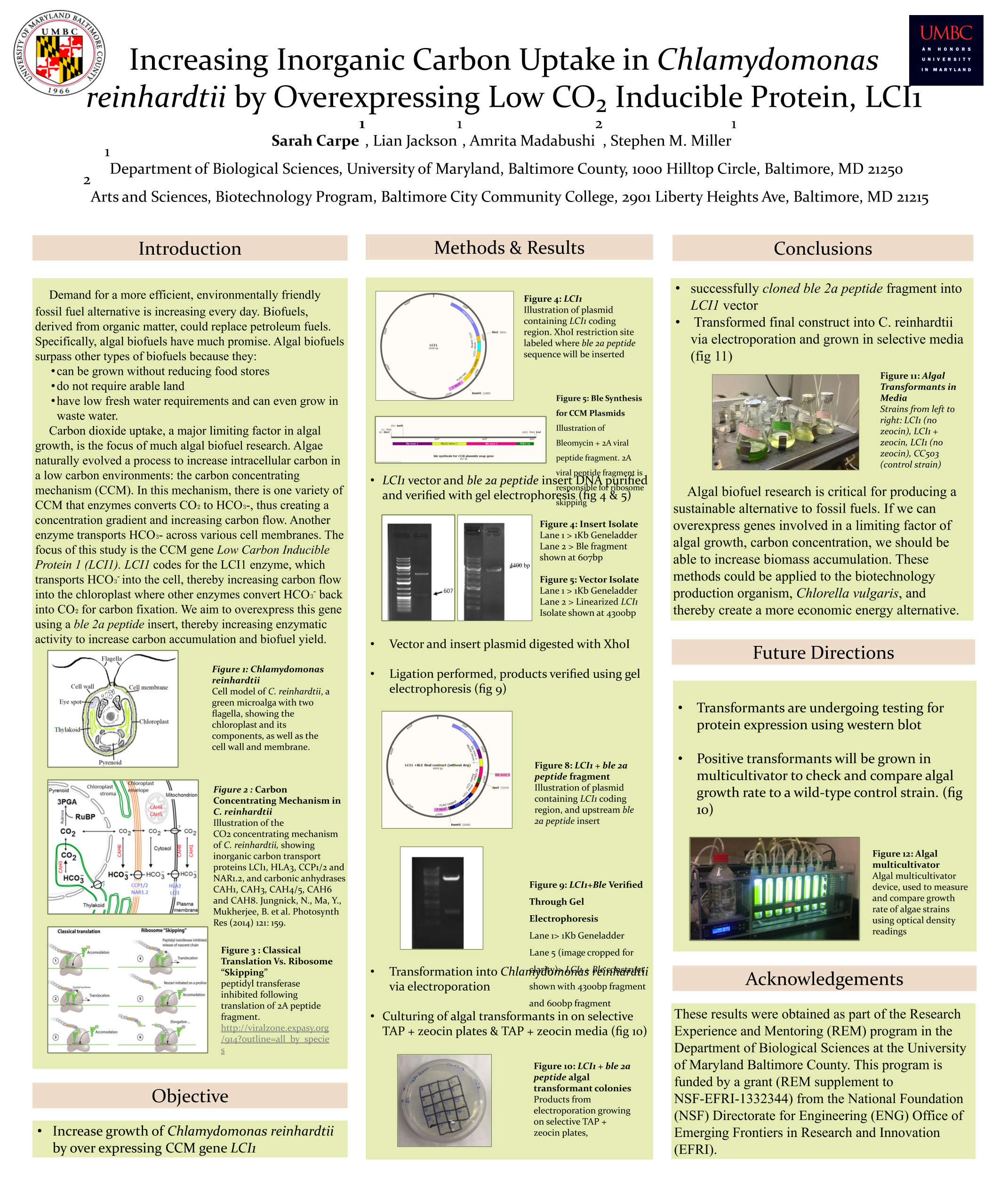
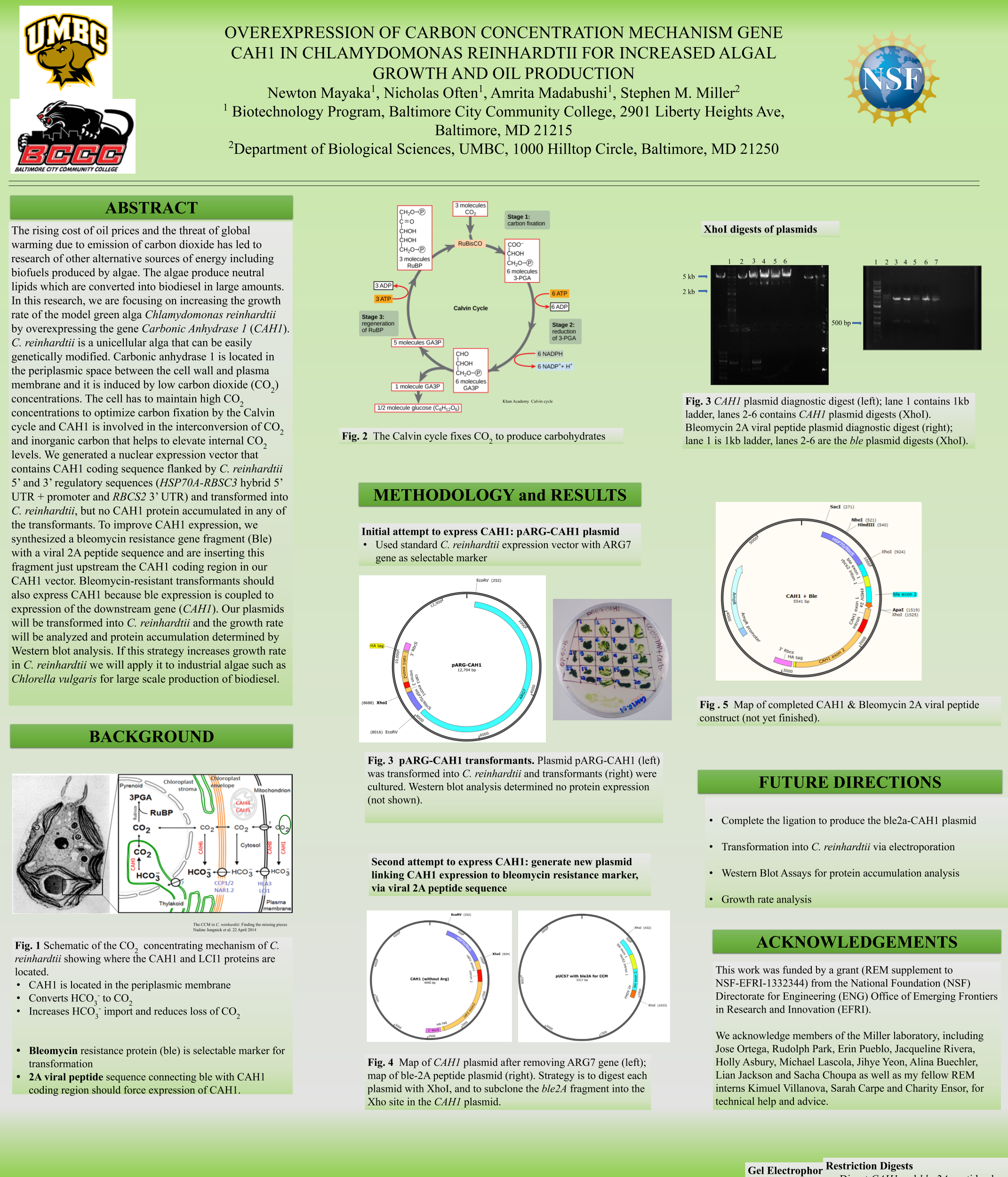
Biofuels are obtained from biomass and are a source of renewable energy that is more sustainable than fuels derived from petroleum. We aim to make algal based biofuels more efficient by increasing the growth rate and lipid production in algae. Algae are limited for uptake of carbon dioxide (CO2), which has limited solubility in water. We hypothesize that certain enzymes that concentrate CO2 in algae are limiting for growth, and we predict that overexpressing them will speed growth and biomass production. Chlamydomonas reinhardtii, a green microalga, is a model organism that can be genetically manipulated to overexpress certain proteins. This investigation focuses on CAH3, a carbonic anhydrase that catalyzes the conversion of carbonic acid into CO2, increasing its concentration in the chloroplast pyrenoid, where it is fixed into sugars by the enzyme rubisco. We first designed and ordered a synthetic CAH3 coding region (with HA epitope tag sequence at 3’ end) flanked by XhoI and BamHI restriction sites at its 5’ and 3’ ends. Respectively. Then we used restriction digests to isolate this gene fragment, and we used DNA ligase to clone it into our C. reinhardtii expression vector (pUC-ARG) to produce plasmid pUC-ARG-CAH3. After transforming this construct into C. reinhardtii strain CC4350, we used western blot analysis with anti-HA antibody to test for CAH3 abundance, but none of the transformants we tested had detectable levels of CAH3 protein. In an attempt to improve expression, we next replaced the ARG7 selectable marker gene with a bleomycin resistance cassette (ble-2A) that couples the expression of the ble protein (through a viral 2a-peptide sequence) to the CAH3 sequence. To this end we digested pUC-ARG-CAH3 with EcoRV to remove the ARG7 gene, religated, then digested with XhoI and ligated a gene-synthesized ble-2A fragment flanked with XhoI sites into the expression vector. Restriction analysis and sequencing of plasmid minipreps confirmed that one ligation product had the fragment cloned in correct orientation. We are now using electroporation to transform this plasmid into C. reinhardtii, then we will use western blot analysis to test transformants for protein expression. If transformants are obtained that overexpress CAH3, we will use an algal multicultivator to compare their growth rate to the wild type using the muti-cultivator. Consequently, if increased growth is achieved, the same procedure can be applied to the industrial alga Chlorella vulgaris for production of biofuels.